Liquid Biopsy (LB) Core
The liquid biopsy core can perform the isolation of circulating markers such as rare cells (for example, circulating tumor cells, CTCs), cell free molecules (such as circulating cell free DNA), and extracellular vesicles (EV) in highly efficient and reproducible ways with full automation. The Core can isolate LB markers from a variety of clinical samples including blood, urine, sweat, saliva, and cerebrospinal fluid as well as others. Following the isolation, the amount (number of cells or EV particles or the mass of cell free molecules) of enriched LB marker can be counted and the marker subjected to a variety of molecular assays, such as droplet digital PCR, RT-qPCR, Western blotting, and Next Generation Sequencing. However, the isolate can be returned to the user for molecular processing in his/her own laboratory. A robotic platform can accept samples in standard tubes, such as blood EDTA tubes, and fully automate the isolation using unique technology specific to the Core. Blood samples can be seeded with a stabilizer cocktail to allow shipment of samples at room temperature to the Core (stability for up to 72 h post-draw).
Due to the robotic nature of the isolation process, the results are highly reproducible and the Core can perform 36 isolations per day. The Core has a specially designed microfluidic chip for optimal performance in terms of recovery and purity for the isolation of rare cells, cell free molecules, and EVs. The same robot can process a clinical sample for any of the LB markers by selecting the appropriate microfluidic chip.

We can process samples for internal and external users (academic and private). RATES for isolation of Rare Cells, EVs, or cfDNA are $75 (KU-L and KUMC users), $145 for external academic clients, and $225 for private sector clients. If you require enumeration of the isolated markers, additional charges will incur. This can be discussed on a case-per-case basis with the cost dependent on the marker to enumerate (CTCs versus EVs) and the assays that are performed for the enumeration. We can determine cfDNA content as well from the isolate.
Other equipment available to the Core:
- BioRad droplet digital PCR
- Agilent Tapestation
- Illumina Next Generation Sequencer (DNA and RNA-Seq)
- Fluorescence microscopes for immunophenotyping cells
- Transmission Electron Microscope (TEM)
- Nanoparticle Tracking Analysis (NTA)
- Western Blotting
- Ultracentrifuge
- Benchtop centrifuges
- RT-qPCR
- Nanodrop UV/vis
- PCR machines
- Scanning Electron Microscope (SEM)
- -80° C Freezer for sample storage
- Cellometer
- Several brightfield microscopes
Rare Cell Isolation
We use a plastic chip to affinity isolate the biological cells of choice by the user. The plastic chip can be tailored for any antibody or other affinity agent, such as aptamers to target the necessary cells. The chip has been demonstrated to efficiently isolate CTCs, circulating leukemia cells, circulating multiple myeloma cells, and leukocyte subsets. The recovery is high (>95% for high expression of the target antigen in the cell). Superb purity (>85%). Because of the high purity, enriched cells can be released from the device and submitted to molecular processing in a batch mode (i.e., no single cell picking is required).

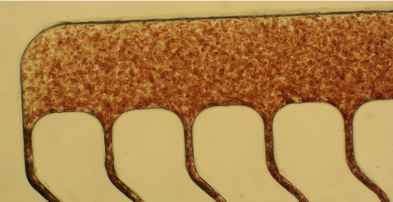
The chip can process 3 mL of clinical sample in approximately 45 min. The chip can be operated by the fluid handling robot in the Core to fully automate the sample processing. Cells can be released from the chip, spun onto a slide using a cytospin, stained, and the cells phenotyped using a highly sensitive fluorescence microscope. The Core also has a CellSearch instrument that can be used for CTC isolation.

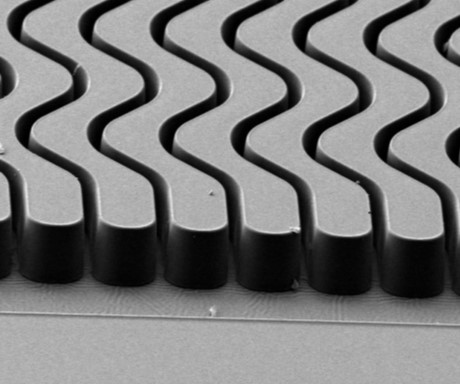
Rare cell affinity isolation chip. The chip is made from a plastic via injection molding and consists of a series of sinusoidally-shaped channels. Each channel contains affinity agents that can be tailored to fit the user’s application. This chip can be operated by the robotic platform.
Extracellular Vesicle (EV) Isolation
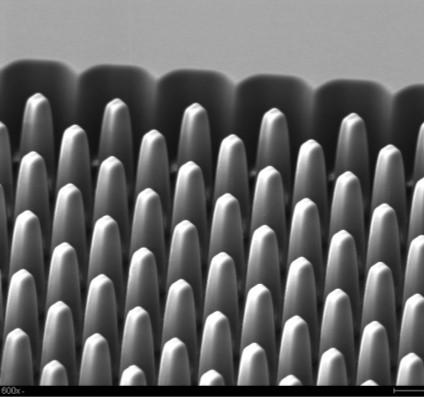
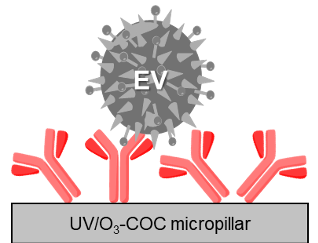
The Core uses a plastic chip consisting of a high density array of micropillars (each chip has 1.5M pillars) that can be activated to allow for the covalent attachment of affinity agents to each pillar. The high surface area of the chip allows for selecting >1011 particles, and release the EVs in a total volume of 22.4 µ L.

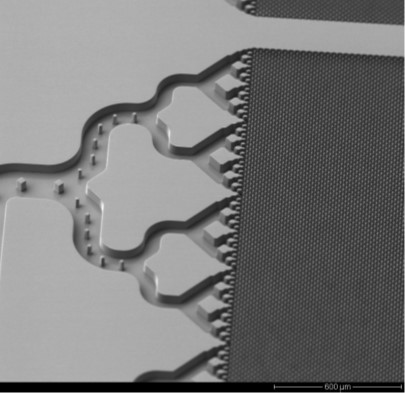
The chip is manufactured via injection molding in a high production mode. The chip can recover >80% of the EVs at a volume flow are ~25 µ L/min. The chip can be operated on the fluid handling robot.
- EVs isolated directly from plasma/serum/urine based on affinity interaction of antigens loaded onto the surface of pillars.
- Can be customized to any surface antigen of interest
- EVs can be released intact from chip for downstream applications
Cell Free DNA (cfDNA), genomic DNA and RNA Isolation
We have developed an inexpensive microfluidic chip that can be used for the solid-phase extraction of DNA (cfDNA, plasmid DNA, genomic DNA) with high efficiency and using a very simple procedure. The chip is made from a plastic that can be injection molded to produce the chip at low cost and in high numbers to support any clinical study. When compared to existing solid-phase extraction techniques (Qiagen, Norgen), it exceeds their recovery values across a large DNA size range (recovery >85%). In addition, the solid-phase extraction chip can be used to extract total RNA as well. The chip can be operated by our fluid handling robot and process 16 samples at any one time and with full automation. Here are the salient features of this solid-phase extraction technique:
- Solid-phase extracted DNA size can be controlled by the immobilization buffer composition (PEG, NaCl, EtOH)
- cfDNA mass load ~2,000 ng per chip
- cfDNA eluted in a small volume, ~20 µL
- Elution buffer of your choice: PCR buffer, water, TE, etc
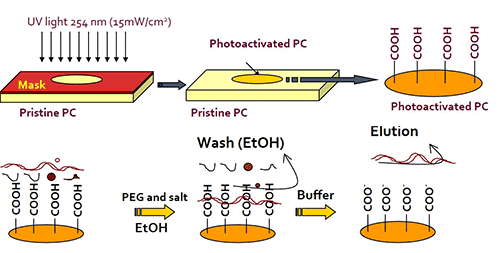
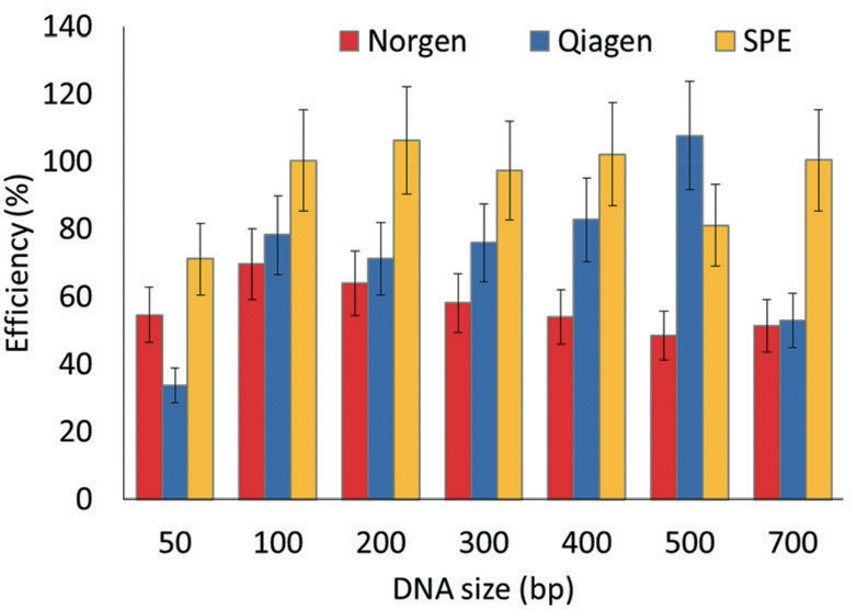
Solid-phase extraction chip for DNA and RNA. The chip is made from a plastic and can accommodate up to 2,000 ng of cfDNA. The chip consists of micropillars to increase provide the high mass load of DNA. By simply changing the composition of the immobilization buffer, which consists of polyethylene glycol (PEG), salt (NaCl or MgCl2), and ethanol (EtOH), the size of DNA preferentially extracted can be selected. Because the chip is made from a plastic, it can be injected molded at very low cost and at high rates. Following molding, the chip requires UV/O3 activation and cover plate bonding and is ready to use for solid-phase extractions.